A Predator Prey Model With Evolution in Agents.jl
Using Julia's Agents.jl to model predator-prey populations adapting to each other
I’ve always been fascinated with the dynamics of evolution. It is incredible to me how predators and prey evolve together in order to sustain themselves. Unfortunately, coming by experimental data of real-time evolution is difficult, so we must resort to some kind of artificial simulations.
To do so, one might recall predator-prey models from their introductory differential equations class. Despite being toy examples, they are a perfect example of how real world phenomena can be distilled to very simple concepts and still provide a lot of insight when modeled properly.
Even though simple models have been proven to answer a lot of questions, they can’t do everything. To answer harder questions that require more variables—such as ones involving adaptations to other organisms—we need to expand our models. To work from the ground up, we will start off with the most basic predator prey model and work our way up to a model that can give us insight into a population of organisms adapting to each other over time.
In the end, I summarize the findings and emphasize some unexpected discoveries in the way our model evolves—make sure to read conclusion for my analysis of the results.
Introduction to the Lotka-Volterra Equations
The classic Lotka-Volterra equations are a wonderful way to represent interaction between predator prey populations:
The familiar equations describe the growth rate of a population of sheep—the prey—and wolves—the predators. It is clear from the equations that the assumptions the model makes is that more prey, or a higher `x`, increases the growth rate of both the prey and the predators, while more predators, a higher `y`, decreases the growth rate of the prey and can decrease the growth rate of the predators. Here is an example of the solution to the ODEs:

Each dot represents a different initial starting value for the number of predators and prey, and the orbits represent how the populations change over time according to the ODEs.
While the assumptions may be simplistic, the resulting dynamics (which you can dive into on the Wikipedia page) can actually be found in some ecosystems and even economic models.
An Example of a Simple Extension to the Lotka-Volterra Equations
Still, we can further extend the Lotka-Volterra equations to understand more complicated dynamics such as introducing grass, represented by `z`, which would interact with the sheep population as such (the predator ODE would remain unchanged):
This time, I will leave it up to you to figure out what assumptions I made and what the constants mean (hint: it would be quite unnatural for the grass to grow to infinity in the absence of any sheep, there must be a limit; and sheep should only grow if there is sufficient food to sustain the population). If you disagree, by all means, share your ideas in the comments!
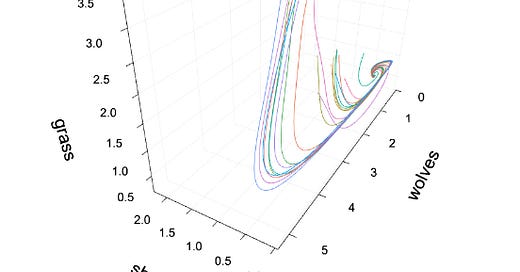
This system of ODEs, too, should yield some interesting dynamics. Using Julia, we can solve the ODEs using a simple Euler solver and obtain the following plot:

For my choice of parameters (all 1’s), it looks like there is an attractive point where all the plotted initial values converge to. If you are able to, I encourage you to run the code for yourself, explore other initial values and parameters, or even test out different forms of the ODEs.
Exploring More Complicated Interactions With Agent Based Modelling
But what if we wanted to explore situations that demand more than the modelling of simple population dynamics? For example, what if we could prescribe a certain poisonous gene to the sheep that wolves can develop a genetic resistance to and see how the genetic pools evolve over time. To be more specific, imagine a sheep-wolf-grass environment that obeys the following rules:
We have a 30 by 30 grid of grass that can be eaten by sheep, and each patch takes 30 days to fully grow after being eaten.
Our sheep are poisonous, and the type of poison the sheep secretes is determined by a gene represented by a number between 0 and 1.
The sheep’s number from 0 to 1 will be referred to as a ‘gene number’
Each wolf is naturally immune to a certain range of toxins. The center of that range is determined by another gene that ranges from 0 to 1 and is found in each wolf.
The wolves’ number from 0 to 1 will be referred to as a ‘gene center’ and their range will be referred to as the ‘gene_range’
When wolves and sheep reproduce, their offspring is a perfect clone of themselves except the offspring’s gene number is given a random mutation.
The gene center, however, will remain constant

With such an environment, it could be interesting to see how the gene distributions run away or chase each other, how mutation and reproduction rates can affect stability, and which populations are are to sustain themselves.
If we want to answer these questions, we need some way to simulate them. However, I certainly cannot think of a system of differential equations which can capture these rules, but—luckily—I don’t have to. Using Julia and Agents.jl, we can employ agent based modelling to actually simulate a population of sheep, wolves, and grass with these rules hard-coded into their behavior.
Due to the popularity of predator-prey models, the Agents.jl package already published an example of discrete predator-prey population dynamics with sheep, wolf, and grass populations. Our job will then be to add in our gene functionality which includes giving wolf and sheep a gene, some way to mutate the gene when reproducing, and only allowing the wolves to eat sheep when they fall within their gene range.
In the next subsection, I will be giving a quick look into how the simulation works and explaining some of the Julia code. Any reader should safely be able to skip the upcoming section if they don’t want to read code or just want to see how such a system evolves over time.
Assigning Genes and Mutation Rates to Agents and Grass
All code can be found in this GitHub repository.
Assigning genes, gene ranges, and mutation rates to the agents (the sheep and the wolves) is as easy as adding new fields to the Sheep and Wolf structs.
@agent Sheep GridAgent{2} begin
energy::Float64 # If energy falls below 0, the Sheep dies
reproduction_prob::Float64 # Chance of reproducing after each step
Δenergy::Float64 # Energy gained from eating Grass
gene::Float64 # Gene number from 0 to 1
mutation_rate::Float64 # Mutation rate
end
@agent Wolf GridAgent{2} begin
energy::Float64 # If energy falls below 0, the Wolf dies
reproduction_prob::Float64 # Chance of reproduction after each step
Δenergy::Float64 # Energy gained from eating Sheep
gene_center::Float64 # The gene number between 0 and 1 for Wolves
# Each Wolf can only eat if |Sheep.gene-Wolf.gene_center| < Wolf.gene_rage
gene_range::Float64
mutation_rate::Float64 # Mutation rate
hunt_range::Int # Vision range for finding edible Sheep
endThe mutation_rate fields come into play when a new agent is added or when grass spreads. For example, in the following code block that is run every time a sheep reproduces, the cloned sheep copies all the traits of the original sheep except its gene number is the original sheep’s gene number plus a random variable randomly sampled from a normal distribution of mean 0 and standard deviation of mutation_rate
function reproduce!(agent::Sheep, model)
# Halves the energy of the sheep that is reproducing
agent.energy /= 2
# Creates a new sheep with a gene number of whatever the current sheep's
# gene number is plus a random number drawn from a normal
# distribution of mean 0 and std of mutation_rate.
# Every other field is a perfect copy of the current sheep
id = nextid(model)
mutated_gene = clamp(agent.gene + rand(model.rng, Normal(0, agent.mutation_rate)), -1, 1)
offspring = Sheep(id, agent.pos, agent.energy, agent.reproduction_prob, agent.Δenergy, mutated_gene, agent.mutation_rate, agent.hunt_range)
add_agent_pos!(offspring, model)
return
endResults of the Simulations
Sheep and Wolves
Now that we have everything set up, we can begin to ask questions about how the genes evolve over time. Let’s see how the populations and gene distributions of wolves and sheep change over time by graphing histograms of their genes over time (the grass is excluded since it doesn’t have any poisonous genes, but know that it’s population oscillates opposite to that of the sheep).

As someone familiar with the concept of evolution would expect, the wolves’ histogram is seen to be ‘chasing’ the sheep’s histogram. This translates into the gene pool of the wolves moving closer to that of the sheep and the sheep’s gene pool moving further away from that of the wolves. Of course, this is explained by the wolves eating an sheep within its gene pool and leaving the ones outside to reproduce in relative safety.
We can visualize chase by graphing the mean gene values for both sheep and wolves over time. By doing so, we also notice that it their ‘seasons’ are quite periodic.

If we increase the wolves’ mutation rate, the red line is able to keep up very well with the sheep’s distribution:
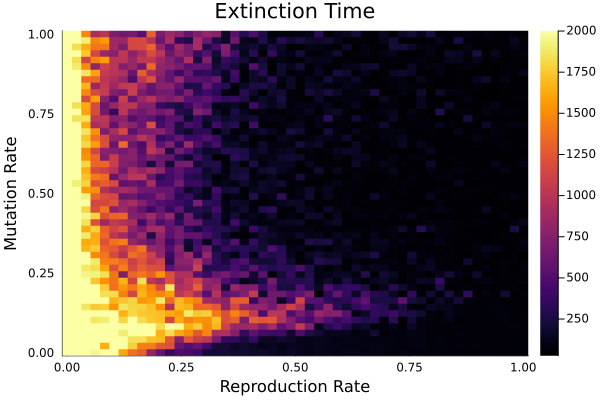
However, the only reason the sheep are able to stay alive is because I set the wolves’ reproduction rate and range of immunity to be very low. Increasing either one quickly killed off all the sheep. If we want to entertain higher wolf reproduction rate or immunity ranges, a lower mutation rate was needed. The following heat map of wolf extinction times might interest some readers:
Sheep and Grass
I left out the grass since they didn’t have any poisonous genes, but what if they did? What if sheep could only eat grass that secretes poison that is similar in gene number to its own poison?
As we did with the sheep and wolf populations, lets first take a look at how their gene distributions change over time:

Surprisingly, they start at their specified initial distributions and then even out over time. Even though such a scenario was never observed with just sheep and wolves, for sheep and grass, this pattern was present for nearly all combinations of initial distributions, reproduction rates, and mutation rates (there were some very rare situations where the sheep died out—try to see if you can make up a scenario which forces the extinction of sheep).
Sheep, Wolves, and Grass
Now, what if we take the next step and try to introduce the wolves into this sheep-grass system? Well, my first 100 attempts at finding a stable system looked like this:
Trying to find what kinds of wolf populations could be sustained by the environment was no easy task. If we don’t consider the initial population numbers, initial distributions, energy gained from eating, and and hunting ranges, there are a total of 7 variables—all numbers between 0 and 1—that we can control (I will say which variables in a moment, their specifics are not too important right now). That means we would need to search in a seven dimensional grid for stable conditions. With just two organisms exhibiting poisonous adaptations (sheep and grass or sheep and wolves), we were lucky enough to find that nearly any initial configuration was stable, but finding the right conditions for the wolf population be able to sustain itself was much more difficult.
In most cases, the wolves would quickly rise in numbers no matter how small the initial population was, eat all the sheep, and then starve themselves to death—leaving only a lush 900 patches of grass. Another common scenario was that the wolves nearly deplete all the sheep, lose most of their population due to the low sheep population, and then not reproduce fast enough to replace themselves before they die of exhaustion (if a wolf’s energy level dips below 0 it dies, and it takes 1 energy to move). The sheep, however, pull through and they happily live on eating grass for the rest of the simulation.
Brute forcing through the 7-dimensional state space to find stable configurations would take forever, which is why I wrote a simple Particle Swarm Optimization (PSO) algorithm to find the combination of variables for which the wolf population is able to stay alive for the longest. I will not be going into how the PSO algorithm works here, but curious readers can reference sections 16.1.1-16.1.5 of these notes for a good geometric intuition or the Wikipedia page for some fast facts. For our purposes, you just need to understand that some n particles collectively search and adjust their trajectory towards the best positions in the 7 dimensional space.
During the optimization process, we also generate some data to play around with—in our specific implementation, we had 20 particles in the swarm and each particle made 100 searches, so 2000 data-points in total. This means the PSO can be useful in two ways: finding a good set of parameters for a stable wolf population and allowing us to explore, in a very loose sense, the regions of our seven dimensional space which are most likely to produce stable populations. The latter was of particular interest to me since I wanted to know what sets stable configurations apart.
Running the PSO, we can plot out various 2d relationships in a pairwise plot to see if we can find any regions of stability. In the following plot, redder regions correspond to those positions that the PSO algorithm preferred to find stable configurations in. If we allow all parameters to range between 0.1 and 1 (while technically values of 0 are allowed, configurations where, say, wolf populations that just don’t grow at all over time won’t be very interesting; and, indeed, when I do allow PSO to pick values of 0, it sets as many variables to 0 as soon as possible), the regions PSO explored can be plotted as follows (and for the long-awaited reveal of the 7 variables):

Hey, I never said it would look pretty. When looking at this pairwise plot, you should keep in mind that the red regions do not exactly correspond to the regions where stable configurations are the most likely (I defined a stable configuration as one where to wolves survive for at least 2000 time-steps). Instead there is a subtle difference: the red regions is where the PSO algorithm most preferred to look for stable solutions. We can distill the findings from the pairwise plot into a few observations about the PSO’s preferences.
The PSO algorithm preferred looking in regions where
sheep_reproducehas a value less than 0.3wolf_reproduce,wolf_mutation_rate,grass_gene_range, andwolf_gene_rangeare all extremely low—all very near to 0.1 by the looks of it.sheep_mutation_ratewas above 0.5. But this variable had some more freedom since we can see a few times when stable configurations were found at lower values.
As for two dimensional relationships, we also have
lower
sheep_mutation_raterates correspond with slightly lowersheep_reproducevaluesas
grass_mutation_ratesdecrease, the PSO algorithm converges to asheep_reproducevalue of ~0.15.
To point out just how delicate this system is. When I change the minimum value of the variables to be 0.2 rather than 0.1, the PSO algorithm is completely lost. The best run it had only sustained wolves for about 450 time-steps and the pairwise plot (literally) pales in comparison. The few iterations that produced stable outputs in the above pairwise plot were all very uneventful as seen in the following animation

We see that the grass-sheep interaction dominates the evolution as they even out in gene distributions, but the wolf population hardly interacts with the sheep. The only stable configurations that PSO found seem to all diminish the effects of the wolves. Clearly, in our environment, a three body system of genes is very unsustainable. Gene interactions had to be limited to being between only two organisms at a time in order for a stable ecosystem.
Concluding Thoughts and Summary
As I hoped, modelling poisonous organisms allowed to me to view evolutionary mechanisms that push predator-prey pairs to continually try and eat and evade one another, respectively. It was interesting to find that the model did not present any equilibrium states where both the sheep and wolf gene distributions stayed constant, I wonder if organisms in nature are also continuously evolving instead of finding equilibrium. It’s hard for me to tell because I’m no biologist and also I’ve never observed evolution in real time, so I sadly have don’t have an intuitive grasp on how it works.
As to why the wolves’ distribution can be seen to chase that of the sheep, but the grass’ distribution evens out over time if given a poison trait, I have no good explanation! If someone can provide a testable idea for me to try out, I would love to fill in this gap with a working theory.
Even though I could not find stable three organism dynamics, something interesting to note about this whole experiment is that the basis of the ‘genes’ was some kind of poison secretion/adaptation; while there are plenty of examples of organisms developing resistance to toxins in order to access a food source, I cannot seem to find any examples of three different organisms in different trophic levels whose poisonous adaptations all interact with one another like in the sheep-wolf-grass system (for example, I want to find something like a poisonous frog being eaten by immune predators, and, at the same time, eating insects that give off the same poison).
I am certain that assuming the rarity of three organism with the same type of poison adaptations is explained by this model is a gigantic leap in logic. I openly admit that I have no evidence of any physical relevance of this model! However, even if nothing is true, it was still an interesting exercise in agent based modelling, predator-prey dynamics, visualizations, and also playing around with the parameters taught me about some of the the complex interactions that ecosystems exhibit—not nearly as intuitive as the functions I am used to dealing with as a physics and mathematics student!
Some next steps that I (or maybe a motivated reader who I would be very happy to respond to if they reached out) might try to implement are:
Pack/herding behavior for the wolves and sheep to find more optimal ways to hunt prey/evade predators
More complex boundaries such as narrow passage ways and mountains to observe genetic drift due to geographic isolation
Have more than one gene (such as poison as well as camouflage) which I conjecture should allow for more than two organisms’ genes to interact at once
External factors such as viruses or weather
This is my first post on Substack, and I hope to keep this updated with more projects (and maybe puzzles) in physics, mathematics, and computational sciences. If you enjoyed what you read or have feedback, let me know!